4. Needleman-Wunsch (NW)¶
Catalog of Features¶
In this section, you will learn about the following components in Spatial:
- FSM
- Branching
- FIFO
- Systolic Arrays
- File IO and text management
- Asserts, Breakpoints, and Sleep
Application Overview¶
The Needleman-Wunsch algorithm is an algorithm used in bioinformatics to align protein or nucleotide sequences. It builds a scoring matrix based on two strings, and then backtraces through the score matrix to determine the alignment of minimum error. For more information on the algorithm’s history and implementations, visit the Wikipedia page (https://en.wikipedia.org/wiki/Needleman-Wunsch_algorithm). The image below (credit Wikipedia) demonstrates a rough overview of how the algorithm works.
Data Setup and Validation¶
In this algorithm, we will assume that a domain-expert keeps data files of DNA sequences, seqA.txt and seqB.txt.
We will create an app that will read whatever text is in these files, pass it to the FPGA, return with the best alignment
between the two, and print out the result back into files called alignedA.txt and alignedB.txt. In order to test
if our alignments are “correct,” we will aim to have less than 10% of the entries be in error.:
import spatial.dsl._
import virtualized._
object NW extends SpatialApp {
@virtualize
def main() {
val seqA_text = loadCSV1D[MString]("/home/mattfel/seqA.txt", " ").apply(0) // Loads into array with 1 string
val seqB_text = loadCSV1D[MString]("/home/mattfel/seqB.txt", " ").apply(0) // Loads into array with 1 string
println("Aligning " + seqA_text + " and ")
println(" " + seqB_text)
val seqA_data = argon.lang.String.string2num(seqA_text) // Returns array of Int8
val seqB_data = argon.lang.String.string2num(seqB_text) // Returns array of Int8
val seq_length = seqA_data.length
val LEN = ArgIn[Int]
val LEN2x = ArgIn[Int]
setArg(LEN, seq_length)
setArg(LEN2x, seq_length*2)
val seqA = DRAM[Int8](LEN)
val seqB = DRAM[Int8](LEN)
setMem(seqA, seqA_data)
setMem(seqB, seqB_data)
val seqA_aligned = DRAM[Int8](LEN)
val seqB_aligned = DRAM[Int8](LEN2x)
val max_length = 256
val d = argon.lang.String.char2num("-")
val dash = ArgIn[Int8]
setArg(dash,d)
val u = argon.lang.String.char2num("_")
val underscore = ArgIn[Int8]
setArg(underscore,u)
Accel{}
val seqA_aligned_result = getMem(seqA_aligned)
val seqB_aligned_result = getMem(seqB_aligned)
val errors = seqA_aligned_result.zip(seqB_aligned_result){(a,b) => if (a != b && a != d && b != d) 1 else 0}.reduce{_+_}
println("Found " + errors + " errors out of " + {seq_length*2} + " characters")
val cksum = errors.to[Float] / (seq_length*2).to[Float] < 0.1.to[Float]
println("Acceptable? " + cksum)
val seqA_aligned_string = argon.lang.String.num2string(seqA_aligned_result)
val seqB_aligned_string = argon.lang.String.num2string(seqB_aligned_result)
println("Aligned A: " + seqA_aligned_string)
println("Aligned B: " + seqB_aligned_string)
}
}
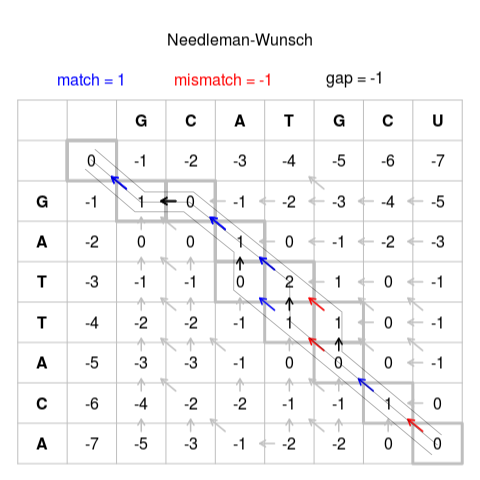
Score Matrix Population¶
In this first section, we will make a forward pass to fill out the score matrix. In this algorithm,
we need to embed two pieces of information in each matrix entry: the score at that entry and the direction
to travel to achieve that score (N, W, or NW). We will start by defining a new struct that can contain
this tuple up above our main():
@struct case class nw_tuple(score: Int16, ptr: Int16)
As we traverse the score matrix, we check the left, top, and top-left entries, add a score update, and check which path gives us the maximum score for that entry. To determine the additional score when coming from the top-left, we check if the letter at the top of the column (from string A) matches the letter from the left of the row (from string B). If there is a match, then this path is rewarded with an addition of 1. If they do not match, then we penalize this path with a score of -1. We then look at the cost when coming from the left and from the top. These transitions correspond to skipping an entry in B and skipping an entry in A, respectively, and we penalize them as they do not correspond to string matches. This transition is called a “gap.” Let’s now assign vals to keep track of these properties:
val SKIPB = 0 // move left
val SKIPA = 1 // move up
val ALIGN = 2 // move diagonal
val MATCH_SCORE = 1
val MISMATCH_SCORE = -1
val GAP_SCORE = -1
Now, we can write the code that will traverse the matrix from top-left to bottom-right and update each entry
of the score matrix. Note that along the left edge and the top edge of the score matrix, we initialize the
scores by -1 each for each hop away from the top left corner. Then, for each entry, we first compute if there is a match
between the elements in string A and string B. We then proceed to compute the from_left, from_top, and from_diag
updates based on these values and choose the smallest of them. When getting this result, we keep the tuple that consists of
both the new score and the path taken to achieve this new score. Finally, we update the score matrix so that this new
value is available for the next update:
Accel{
val seqa_sram_raw = SRAM[Int8](max_length)
val seqb_sram_raw = SRAM[Int8](max_length)
seqa_sram_raw load seqA(0::LEN)
seqb_sram_raw load seqB(0::LEN)
val score_matrix = SRAM[nw_tuple](max_length+1,max_length+1)
Foreach(LEN+1 by 1){ r =>
Sequential.Foreach(0 until LEN+1 by 1) { c =>
val previous_result = Reg[nw_tuple]
val update = if (r == 0) (nw_tuple(-c.as[Int16], 0)) else if (c == 0) (nw_tuple(-r.as[Int16], 1)) else {
val match_score = mux(seqa_sram_raw(c-1) == seqb_sram_raw(r-1), MATCH_SCORE.to[Int16], MISMATCH_SCORE.to[Int16])
val from_top = score_matrix(r-1, c).score + GAP_SCORE
val from_left = previous_result.score + GAP_SCORE
val from_diag = score_matrix(r-1, c-1).score + match_score
mux(from_left >= from_top && from_left >= from_diag, nw_tuple(from_left, SKIPB), mux(from_top >= from_diag, nw_tuple(from_top,SKIPA), nw_tuple(from_diag, ALIGN)))
}
previous_result := update
if (c >= 0) {score_matrix(r,c) = update}
}
}
}
While it is possible to parallelize the row updates in this algorithm, it is a little tricky because you should not update any entry until you have all of its three adjacent source entries. See (TODO: link to spatial-apps) for an example on how to safely parallelize across rows.
Score Matrix Traceback¶
Now we can traverse the score matrix, starting from the bottom right. We will use a FIFO to store the aligned result, and a finite state machine (FSM) to handle the back trace and complete when the FIFOs are filled. The state in the FSM starts at 0, which we use for the state to trace back through the matrix. When we either hit the top edge or the left edge of the score matrix, we jump to state 1 which is used to pad both of the FIFOs until they fill up. Once the FSM detects that they are full, it exits and the results are stored to DRAM. The branch conditions in this FSM demonstrate how we can use if/then/else to arbitrarily execute parts of the hardware.:
val traverseState = 0
val padBothState = 1
val doneState = 2
val seqa_fifo_aligned = FIFO[Int8](max_length*2)
val seqb_fifo_aligned = FIFO[Int8](max_length*2)
val b_addr = Reg[Int](0)
val a_addr = Reg[Int](0)
b_addr := LEN
a_addr := LEN
val done_backtrack = Reg[Bit](false)
FSM[Int](state => state != 2) { state =>
if (state == traverseState) {
if (score_matrix(b_addr,a_addr).ptr == ALIGN.to[Int16]) {
seqa_fifo_aligned.enq(seqa_sram_raw(a_addr-1), !done_backtrack)
seqb_fifo_aligned.enq(seqb_sram_raw(b_addr-1), !done_backtrack)
done_backtrack := b_addr == 1.to[Int] || a_addr == 1.to[Int]
b_addr :-= 1
a_addr :-= 1
} else if (score_matrix(b_addr,a_addr).ptr == SKIPA.to[Int16]) {
seqb_fifo_aligned.enq(seqb_sram_raw(b_addr-1), !done_backtrack)
seqa_fifo_aligned.enq(dash, !done_backtrack)
done_backtrack := b_addr == 1.to[Int]
b_addr :-= 1
} else {
seqa_fifo_aligned.enq(seqa_sram_raw(a_addr-1), !done_backtrack)
seqb_fifo_aligned.enq(dash, !done_backtrack)
done_backtrack := a_addr == 1.to[Int]
a_addr :-= 1
}
} else if (state == padBothState) {
seqa_fifo_aligned.enq(underscore, !seqa_fifo_aligned.full)
seqb_fifo_aligned.enq(underscore, !seqb_fifo_aligned.full)
} else {}
} { state =>
mux(state == traverseState && ((b_addr == 0.to[Int]) || (a_addr == 0.to[Int])), padBothState,
mux(seqa_fifo_aligned.full || seqb_fifo_aligned.full, doneState, state))
}
seqA_aligned(0::LEN2x) store seqa_fifo_aligned
seqB_aligned(0::LEN2x) store seqb_fifo_aligned
Generally, an FSM is a hardware version of a while loop. It allows you to arbitrarily branch between control structures and selectively execute code until some breaking state condition is reached.
Asserts, Breakpoints, and Sleep¶
There are sometimes cases where the app writer wants to escape the app early or pause the app for a period of time. In this subsection we will explore how to implement the breakpoint/exit and sleep functions in Spatial.
Firstly, we will discuss breakpoints. These could be for debugging purposes, such as determining why a non-deterministic app is hanging on the FPGA, or for practical purposes, such as handling errors when decompressing a faulty JPEG header. Spatial allows the user to insert breakpoints arbitrarily in the code and will exit the application early and report which breakpoint triggered the exit, if any, at runtime.
In this example, we will demonstrate how to use breakpoints in Spatial by assuming the app writer wants to halt the NW algorithm the first time a character in either string A, string B, or neither is skipped and wants to know which of these conditions caused the exit:
if (score_matrix(b_addr,a_addr).ptr == ALIGN.to[Int16]) {
...
breakpoint()
} else if (score_matrix(b_addr,a_addr).ptr == SKIPA.to[Int16]) {
...
assert(score_matrix(b_addr,a_addr).ptr != SKIPA.to[Int16], "This is an assert example")
} else {
...
exit()
}
Note that “breakpoint()” in this case is not the same as a breakpoint in software. A breakpoint here causes the entire app to quit, rather than allowing the user to step through code manually. While functionality to switch from the FPGA’s built in clock to a manual clock to let the user manually step through cycles may be implemented in the future, there are no current plans to support this.
The above code may generate output that looks like this if the second breakpoint were reached first (breakpoints are 0-indexed):
===================
Breakpoint 1 triggered!
tutorial.scala:100:23 - This is an assert example
===================
In apps that interact with real external systems, such as pixel buffers, audio devices, and sensors, it may be very useful to make the FPGA stall for a period of time so that it interacts properly with these systems. It can also be useful in debugging, to slow down the speed at which a piece of code executes. While grad students may not get much sleep, Spatial makes it easy to put your FPGA to sleep:
sleep(1000000) // Sleep for ~1000000 cycles, or 8ms for a 125MHz clock
Final Code¶
Here is the final code for this version of NW:
import spatial.dsl._
import virtualized._
object NW extends SpatialApp {
@struct case class nw_tuple(score: Int16, ptr: Int16)
@virtualize
def main() {
val SKIPB = 0 // move left
val SKIPA = 1 // move up
val ALIGN = 2 // move diagonal
val MATCH_SCORE = 1
val MISMATCH_SCORE = -1
val GAP_SCORE = -1
val seqA_text = loadCSV1D[MString]("/home/ChrisWunsch/seqA.txt", " ").apply(0) // Loads into array with 1 string
val seqB_text = loadCSV1D[MString]("/home/ChrisWunsch/seqB.txt", " ").apply(0) // Loads into array with 1 string
println("Aligning " + seqA_text + " and ")
println(" " + seqB_text)
val seqA_data = argon.lang.String.string2num(seqA_text) // Returns array of Int8
val seqB_data = argon.lang.String.string2num(seqB_text) // Returns array of Int8
val seq_length = seqA_data.length
val LEN = ArgIn[Int]
val LEN2x = ArgIn[Int]
setArg(LEN, seq_length)
setArg(LEN2x, seq_length*2)
val seqA = DRAM[Int8](LEN)
val seqB = DRAM[Int8](LEN)
setMem(seqA, seqA_data)
setMem(seqB, seqB_data)
val seqA_aligned = DRAM[Int8](LEN)
val seqB_aligned = DRAM[Int8](LEN2x)
val max_length = 256
val d = argon.lang.String.char2num("-")
val dash = ArgIn[Int8]
setArg(dash,d)
val u = argon.lang.String.char2num("_")
val underscore = ArgIn[Int8]
setArg(underscore,u)
Accel{
val seqa_sram_raw = SRAM[Int8](max_length)
val seqb_sram_raw = SRAM[Int8](max_length)
seqa_sram_raw load seqA(0::LEN)
seqb_sram_raw load seqB(0::LEN)
val score_matrix = SRAM[nw_tuple](max_length+1,max_length+1)
Foreach(LEN+1 by 1){ r =>
Sequential.Foreach(0 until LEN+1 by 1) { c =>
val previous_result = Reg[nw_tuple]
val update = if (r == 0) (nw_tuple(-c.as[Int16], 0)) else if (c == 0) (nw_tuple(-r.as[Int16], 1)) else {
val match_score = mux(seqa_sram_raw(c-1) == seqb_sram_raw(r-1), MATCH_SCORE.to[Int16], MISMATCH_SCORE.to[Int16])
val from_top = score_matrix(r-1, c).score + GAP_SCORE
val from_left = previous_result.score + GAP_SCORE
val from_diag = score_matrix(r-1, c-1).score + match_score
mux(from_left >= from_top && from_left >= from_diag, nw_tuple(from_left, SKIPB), mux(from_top >= from_diag, nw_tuple(from_top,SKIPA), nw_tuple(from_diag, ALIGN)))
}
previous_result := update
if (c >= 0) {score_matrix(r,c) = update}
}
}
val traverseState = 0
val padBothState = 1
val doneState = 2
val seqa_fifo_aligned = FIFO[Int8](max_length*2)
val seqb_fifo_aligned = FIFO[Int8](max_length*2)
val b_addr = Reg[Int](0)
val a_addr = Reg[Int](0)
b_addr := LEN
a_addr := LEN
val done_backtrack = Reg[Bit](false)
FSM[Int](state => state != 2) { state =>
if (state == traverseState) {
if (score_matrix(b_addr,a_addr).ptr == ALIGN.to[Int16]) {
seqa_fifo_aligned.enq(seqa_sram_raw(a_addr-1), !done_backtrack)
seqb_fifo_aligned.enq(seqb_sram_raw(b_addr-1), !done_backtrack)
done_backtrack := b_addr == 1.to[Int] || a_addr == 1.to[Int]
b_addr :-= 1
a_addr :-= 1
} else if (score_matrix(b_addr,a_addr).ptr == SKIPA.to[Int16]) {
seqb_fifo_aligned.enq(seqb_sram_raw(b_addr-1), !done_backtrack)
seqa_fifo_aligned.enq(dash, !done_backtrack)
done_backtrack := b_addr == 1.to[Int]
b_addr :-= 1
} else {
seqa_fifo_aligned.enq(seqa_sram_raw(a_addr-1), !done_backtrack)
seqb_fifo_aligned.enq(dash, !done_backtrack)
done_backtrack := a_addr == 1.to[Int]
a_addr :-= 1
}
} else if (state == padBothState) {
seqa_fifo_aligned.enq(underscore, !seqa_fifo_aligned.full)
seqb_fifo_aligned.enq(underscore, !seqb_fifo_aligned.full)
} else {}
} { state =>
mux(state == traverseState && ((b_addr == 0.to[Int]) || (a_addr == 0.to[Int])), padBothState,
mux(seqa_fifo_aligned.full || seqb_fifo_aligned.full, doneState, state))
}
seqA_aligned(0::LEN2x) store seqa_fifo_aligned
seqB_aligned(0::LEN2x) store seqb_fifo_aligned
}
val seqA_aligned_result = getMem(seqA_aligned)
val seqB_aligned_result = getMem(seqB_aligned)
val errors = seqA_aligned_result.zip(seqB_aligned_result){(a,b) => if (a != b && a != d && b != d) 1 else 0}.reduce{_+_}
println("Found " + errors + " errors out of " + {seq_length*2} + " characters")
val cksum = errors.to[Float] / (seq_length*2).to[Float] < 0.1.to[Float]
println("Acceptable? " + cksum)
val seqA_aligned_string = argon.lang.String.num2string(seqA_aligned_result)
val seqB_aligned_string = argon.lang.String.num2string(seqB_aligned_result)
println("Aligned A: " + seqA_aligned_string)
println("Aligned B: " + seqB_aligned_string)
}
}